Automatic chromosome detector
Description of biological application
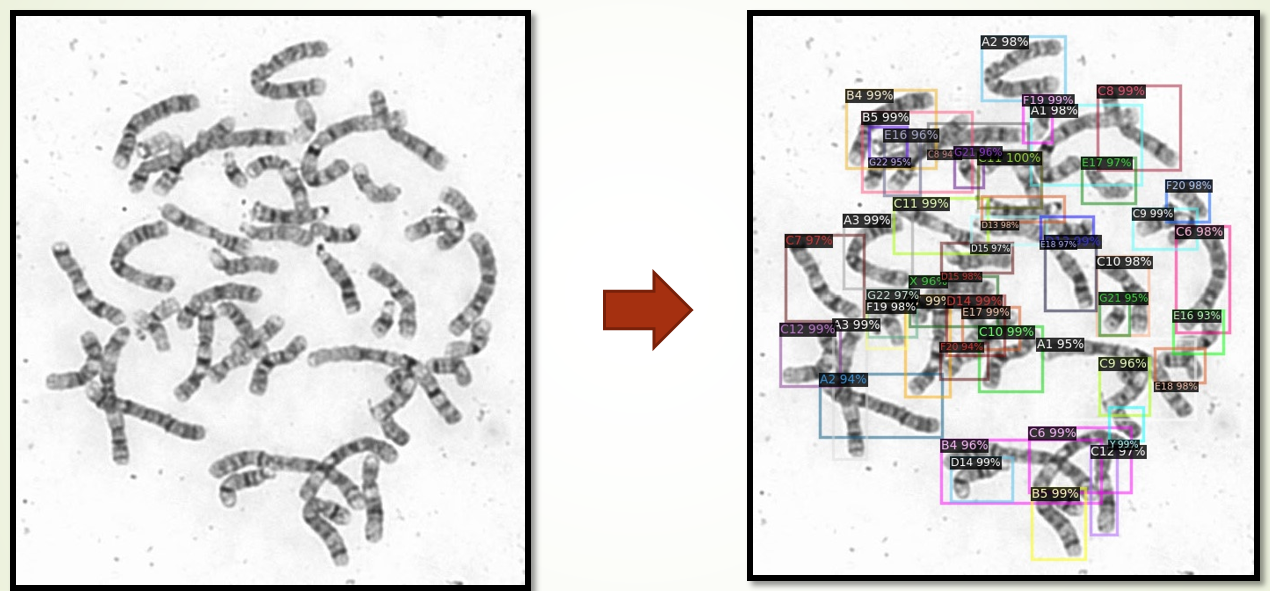
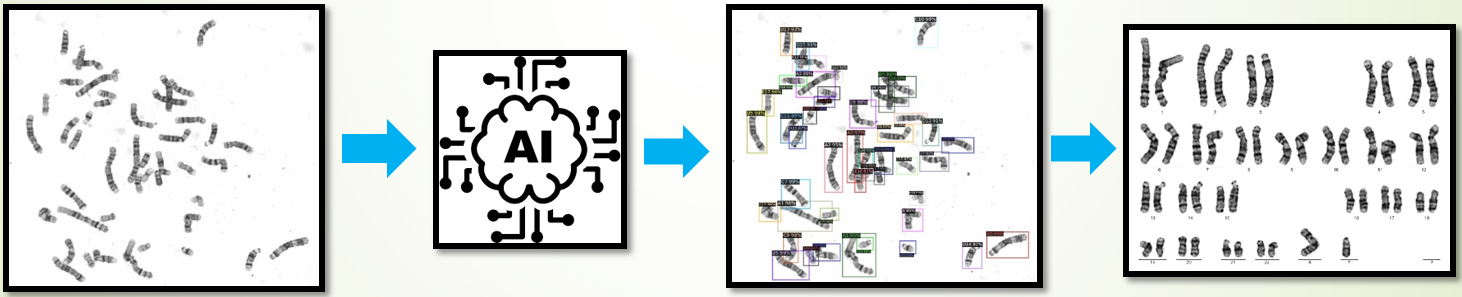
Chromosomes are intracellular aggregates carrying genetic information in genes, which are major objects of study in biological cytogenetics. Chromosome screening is an important part of prenatal care. Manual identification is time consuming and costly (each image takes at least 15 minutes). We have developed an artificial intelligence (AI) model for the automatic chromosome detector based on metaphase cell images using deep learning technology. Moreover, we want to provide the chromosome images and annotations (labels) used to develop the AI model. As far as we know, this is the first publicly available database with the largest number of images and types of labels. The database contains 5,000 metaphase cell images, and each image contains 46 chromosomes (23 pairs). Moreover, the dataset has three different types of annotations: 1). 229,852 object annotations (bounding box) for 24 different chromosomes, 2). 2,000 annotations for a single chromosome, and 3). 5000 pixel-level labels for a single chromosome segmentation. The dataset will be a good performance benchmark for researchers in this field and facilitate the speed and technology development in this application area.
We use in-situ harvest method and trypsin/Wright stain procedure to prepare G-banding. Briefly, the amniotic cells were cultured in BIO-AMF medium (Biological industrial, Beit-Haemek, Israel) for 6 to 8 days.Treating cells with colcemid (NY, USA, Gibco) for 30 min to arrest cells at metaphase. Swell cells with hypotonic solution and fix with methanol/acetic acid mixture. Fixed cells were treated with tryspin then stained with Wright stain solution (MO, Sigma-Aldrich). The karyotype was interpreted according to The International System for Human Cytogenomic Nomenclature (ISCN).
See also: M. J. Barch, T. Kuntsen and J. L. Spurbeck, The AGT Cytogenetics Laboratory Manual, Lippincott-Raven (1997). Yunis J. High resolution of human chromosome. Science 1976: 191:1268-1270. J. McGowan-Jordan, A.
Sim+A13ons and M. Schmid. An International System for Human Cytogenetic Nomenclature. KARGER (2016).
Weight:
Two pre-training models, "best_single_chromosomes.pt" and "best_24_chromosomes.pt",
are provided for training with YOLOv4.
We recommend using argusswift's YOLOv4_pytorch program to operate.
https://github.com/argusswift/YOLOv4-pytorch
| Pretrained model |
Datasets |
mAP50 |
| best_single_chromosomes.ptt |
2000 |
96.5 |
| best_24_chromosomes.pt |
2000 |
90.8 |
Train & test
We have provided the file names for the training and test sets. ("train.txt" "test.txt")
Diffcult image:
The images of the 24 chromosome annotations were evaluated according to the rules, "diff_image.txt" contains the filenames of all difficult images.